Molecular dynamics involved in DNA compaction
Study of Max Planck scientist Vlad Cojocaru on the front cover
DNA is packaged in the cell nucleus by wrapping around proteins called histones to form chromatin fibers. Nucleosomes, the structural units of these fibers, are formed by wrapping 146 base pairs of DNA around eight histones. Additional histone proteins, known as linker histones, bind to nucleosomes in a 1:1 ratio to form chromatosomes. The study of the dynamics of chromatosome formation carried out by Vlad Cojocaru (Max Planck Institute for Molecular Biomedicine, Münster) together with Mehmet Öztürk and Prof. Rebecca Wade of the Heidelberg Institute for Theoretical Studies (HITS) was published in the August issue of Nucleic Acids Research (NAR) with a front cover illustration.
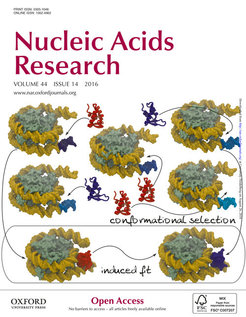
In this study, classical and accelerated molecular dynamics simulations and Brownian dynamics simulations were used to investigate how the linker histone known as H5 binds to nucleosomes. Based on the findings presented, the authors propose that chromatosomes do not have a single defined geometry but multiple ones depending on the dynamic shapes (conformation) of the interacting nucleosome and linker histone. This concept represents an important contribution towards understanding how DNA compacts in the cell nucleus.
The illustration shows, in the upper region, how alternative linker histone conformations (cartoon shapes colored in red and blue) bind to alternative nucleosome conformations (green-yellow surface shapes with a DNA arm in different shades of blue) by a mechanism known as conformational selection. In the lower framed region, an additional mechanism, known as induced fit, is illustrated in which a closed linker histone conformation forms an encounter complex with the nucleosome (left) and converts into an open conformation in the fully bound complex (right). This cover was prepared by Vlad Cojocaru with the molecular visualization program VMD and the graphic editor Inkscape.
HITS
