3D genome organisation independent of gene regulation in Drosophila
How can you pack two metres of DNA into the nucleus of a cell and ensure the right genes are expressed, in the right cell at the right time?
Understanding how our genes function when all the genetic information is packaged into the nucleus is a key question for researchers in the Developmental and Regulatory Genomics group at the MRC London Institute of Medical Sciences (LMS) and the Max Planck Institute for Molecular Biomedicine in Muenster, Germany. Improving our fundamental understanding of the role of the 3D organisation of the genome and regulation of gene expression will help us to better understand changes that lead to mutations and disease, such as cancer.
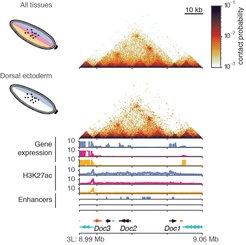
Previous studies have shown genome organisation to be closely linked to gene expression and gene regulation. Recent research from the Developmental and Regulatory Genomics group showed that the 3D genome structure of the Drosophila embryo was established at the onset of zygotic genome activation, an early embryonic stage when most genes begin to be expressed and the embryo begins to form cells. This stage in the Drosophila embryo is approximately equivalent to the 4-cell stage in human embryo and 2-cell stage in mouse embryo development. However, how the 3D genome is then regulated upon cell-fate specification later in development is unknown.
Using Drosophila embryos at this developmental stage as their model system for this study, the researchers explored the link between 3D genome organisation, gene expression and gene regulation. Specifically, they wanted to know how heterogeneous chromatin conformation is across the embryo and whether developmental transcription factors and gene regulation affect genome organisation. “Even at this early stage of development, the embryo has a complex pattern of morphogen gradients that will ultimately determine which cells form given tissues”, said Liz Ing-Simmons, first author of the work.
The Drosophila embryo has an anterior-posterior axis, and a dorsal-ventral axis. In this study the researchers used the dorsal-ventral patterning system to explore how heterogeneous chromatin conformation is specified across the embryo. The specification of tissues in the embryo is determined by the concentration of a transcription factor, Dorsal (DI), in the nuclei. There is higher concentration of Dorsal protein in the ventral part of the embryo which leads to mesoderm, a lower concentration at the dorsal part which leads to dorsal ectoderm and the medium levels lead to neuroectoderm.
The researchers used three models with mutations in the pathway that controls Dorsal concentration (high, medium, low), provided by collaborators at Stockholm University, Sweden. In principle it was expected that the mutations would lead to embryos with different tissue types, all dorsal ectoderm, all neuroectoderm or all mesoderm. This approach would allow the researchers to see if there were differences in the chromatin conformation in these three tissues.
Firstly, the researchers confirmed that the three mutant models had different chromatin states due to activation of different enhancers and different gene expression. Their next question was do these three tissues have different 3D chromatin organisation? Surprisingly, they found that overall, there were no differences in the 3D genome organisation at tissue-specific genes across the three tissue types they investigated.
To explore this in finer detail the researchers pioneered the Micro-C technique in Drosophila in collaboration with scientists at Princeton University, USA. This technique allowed them to examine chromatin organisation at greater resolution than that achieved through Hi-C, revealing several small domains not visible through Hi-C. Even with the increased resolution of the new datasets, 3D genome organisation looked identical in mutant flies.
Juanma Vaquerizas, senior author said: “The increased level of detail provided by Micro-C is a major breakthrough. It will allow us to examine fine level differences in chromatin organisation between samples and understand how this is affected in disease”.
Overall, the group’s findings demonstrate that a change in chromatin state or gene expression is not enough to alter the 3D genome organisation. These results emphasise the role of the 3D genome as an architectural framework for gene regulation, and a distinct organisation of developmentally regulated and housekeeping genes in the Drosophila genome.
Going forwards the Developmental and Regulatory Genomics group will be examining chromatin organisation in different systems using techniques such as Micro-C, to reveal further insights about 3D genome organisation.
‘Independence of chromatin conformation and gene regulation during Drosophila dorsoventral patterning’ was published on 01 April 2021 in the journal Nature Genetics. Read the full article here. https://dx.doi.org/10.1038/s41588-021-00799-x
Text: Dr. Jenna Stevens-Smith, MRC LMS
